Supporting multiple classifications: a proposed change
Part 2: A proposed change to help with using multiple classifications in QuPath
This post is the second of a two-part series looking at how QuPath can handle multiple classifications to start working more meaningfully with multiplex images.
Part #1 showed this in action with a sample image. Here, we see why it is not straightforward.
The question of how QuPath can be used to handle multiple classifications per objects has come up a few times, including in this recent discussion.
In QuPath v0.1.2, it’s only possible to store a single classification per object, but this classification might be derived from another classification. In principle, it should have been possible to have long lists of derived classifications, and thereby smuggle a lot of information into a single classification… but in reality it didn’t really work properly beyond two levels. And the only built-in use of this involved intensity-based sub-classifications.
On the surface, the shift to support multiple classifications would be a big one. It might eventually be worthwhile, but it would also potentially break a lot of existing functionality (and scripts) – and I’d prefer to keep such major changes limited until they are really necessary, and then preferably make them all in one big update (rather than trickling out more slowly over time).
With that in mind, I wanted to see what is the minimal change that I think could be enough for now.
I have a suggestion, and I’m interested in feedback.
The situation until now
Many measurements shown in QuPath are calculated dynamically based on the objects and classifications present for an image.
For example, if I run Positive cell detection for an annotation in a suitable image, I don’t need to request to see the counts of positive and negative cells – these are immediately given with Num Positive and Num Negative values.
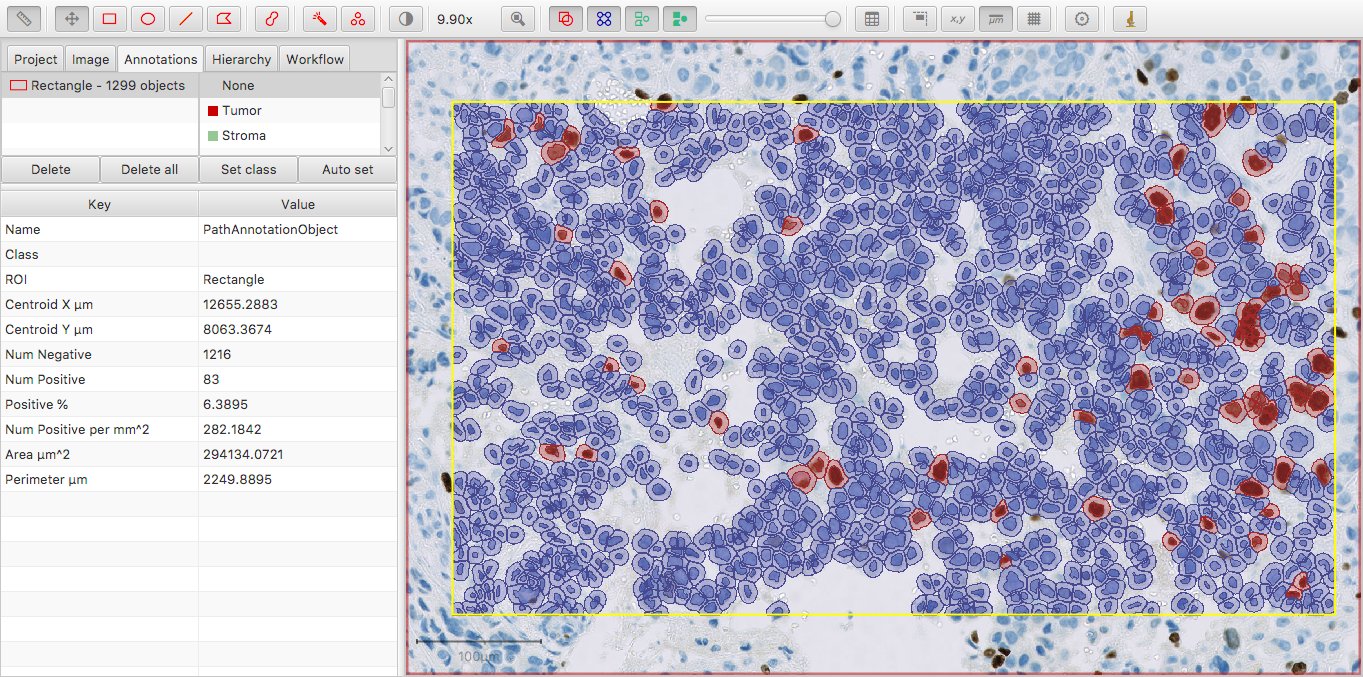
If I then proceed to use Create detection classifier, I can end up with Num Tumor: Positive and Num Tumor: Negative – as well as an overall count of cells classified as tumor under Num Tumor.
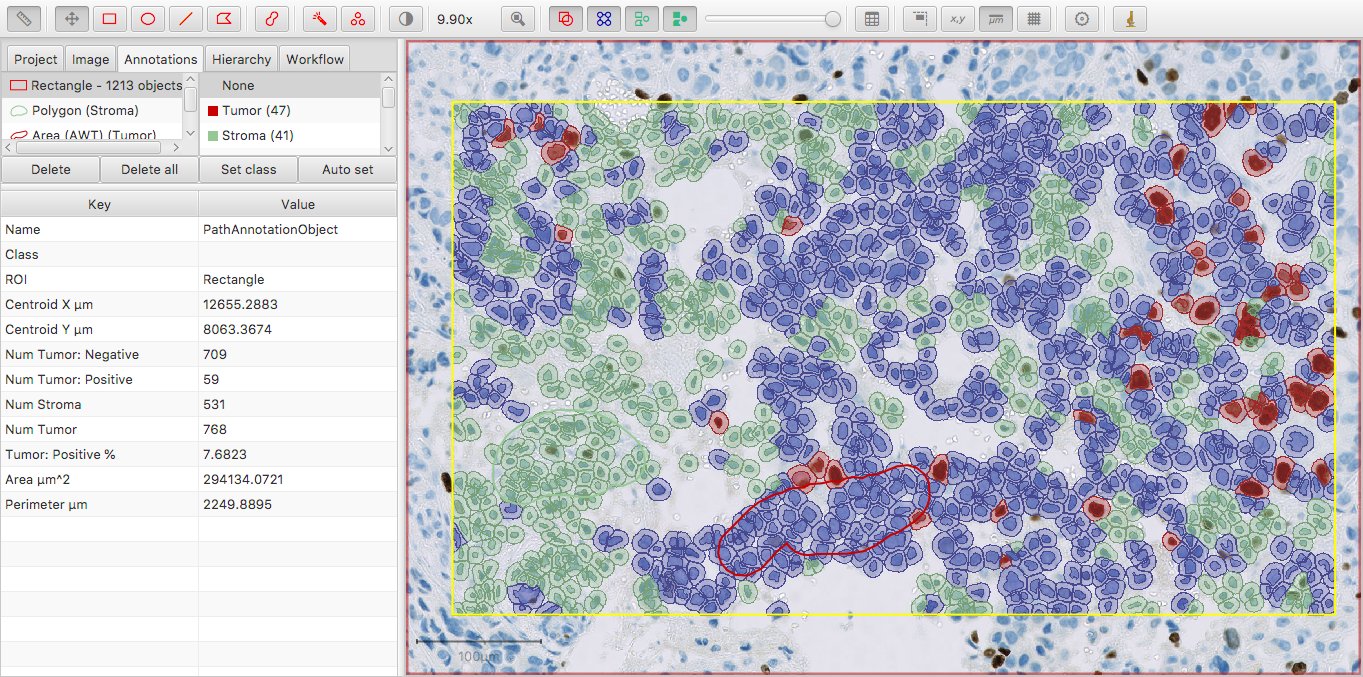
This generally works fine: it’s easy to see that the values in the columns Num Tumor: Positive and Num Tumor: Negative add up to the result in Num Tumor.
However, it’s not necessarily the case that this is so.
For example, someone could run a script that strips the positive/negative classification information from some of the tumor cells:
// Script to remove intensity sub-classification from 10 randomly-chosen 'Tumor' cells
cells = getCellObjects().findAll {it.getPathClass().isDerivedFrom(getPathClass('Tumor'))}
Collections.shuffle(cells)
cells[0..9].each {it.setPathClass(getPathClass('Tumor'))}
fireHierarchyUpdate()
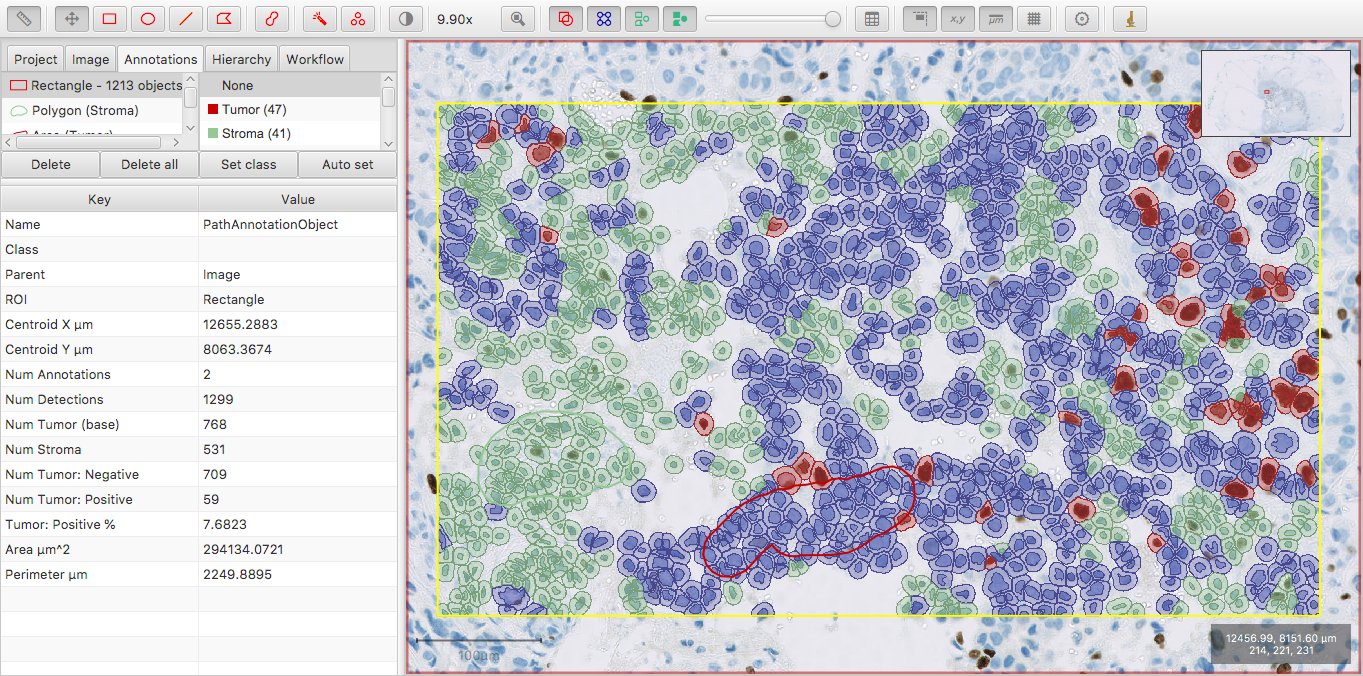
I’ve changed the color of the Tumor class slightly to make it visible in the image; now Num Tumor is 10 more than the sum of Num Tumor: Positive and Num Tumor: Negative.
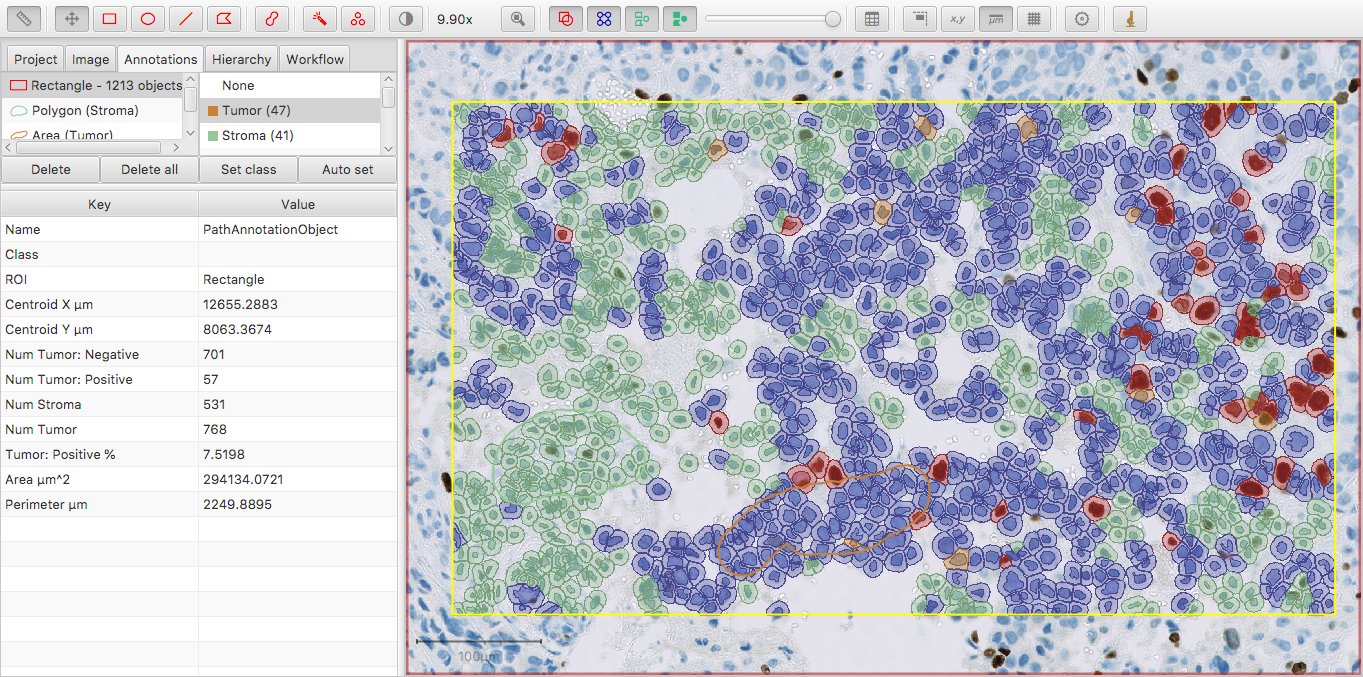
The general principle is this: Num Some Classification gives the count of all cells with exactly Some Classification, or any further classification derived from Some Classification.
The problem with this approach
This is generally ok, and easy to ignore: classifying a type of cell by intensity is generally an all-or-nothing event within QuPath as it stands.
You may justifiably question why anyone would want to run a script like the one above.
However, if we want to reuse the same sub-classification system to represent multiple classifications, this can be confusing.
For example, let’s suppose we have:
- one cell with a classification
CD8 - one cell with a classification
FoxP3 - one cell with a classification
CD8: FoxP3(i.e. it’s ‘double-positive’)
Applying the same ‘dynamic counting logic’ as before, the counts we would expect to see in the annotation table are:
Num CD8 = 2Num FoxP3 = 1Num CD8: FoxP3 = 1
This is depicted (albeit rather artificially) in the following image.
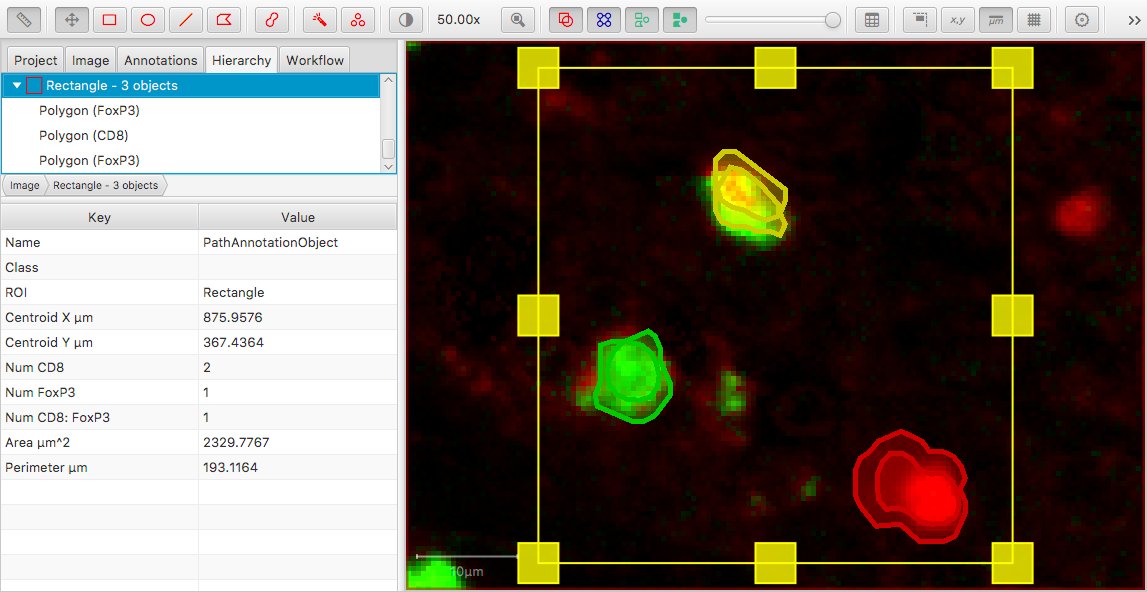
To me, Num CD8 = 2 doesn’t look right and could be surprising; but it happens because we’re counting all objects with classification derived from CD8 – and not just objects with CD8 as the only classification.
On the other hand, we might argue that CD8 = 2 is correct because there are 2 cells classified as CD8 positive. However, then we’d expect to see FoxP3 = 2 as well… but we don’t, because CD8: FoxP3 isn’t derived from FoxP3. The order matters.
I don’t think this is very intuitive, and it is riskily open to misunderstanding.
The proposed change
My proposed change is simple: Num Tumor should give the count of objects with exactly the specified Tumor… and not including any sub-classification derived from it.
Then, in the special case where we are working with intensity-based sub-classifications (i.e. Positive, Negative, 1+, 2+, 3+), a new measurement is created for Num Tumor (base), which is the count of all objects that have the classification Tumor or something derived from Tumor.
Applying this, whenever all our tumor cells are sub-classified by intensity then the results are the same as they were before – except that Num Tumor has been renamed Num Tumor (base).
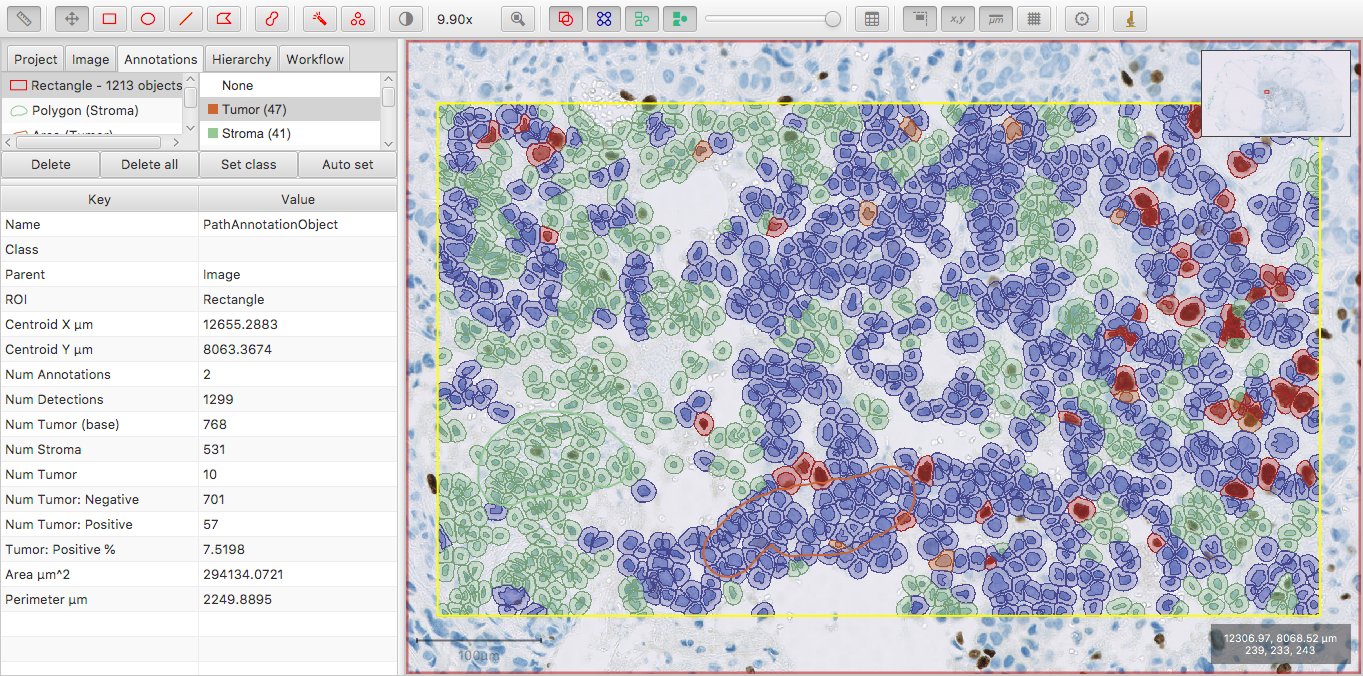
However, whenever we randomly strip the intensity sub-classification from 10 of the tumor cells, then Num Tumor (base) remains the same – but we now get a new Num Tumor measurement of 10, indicating that 10 cells are classified as Tumor only.
Applying this to the CD8 and FoxP3 example gives a different result from before. Now, the columns would include:
Num CD8 = 1Num FoxP3 = 1Num CD8: FoxP3 = 1
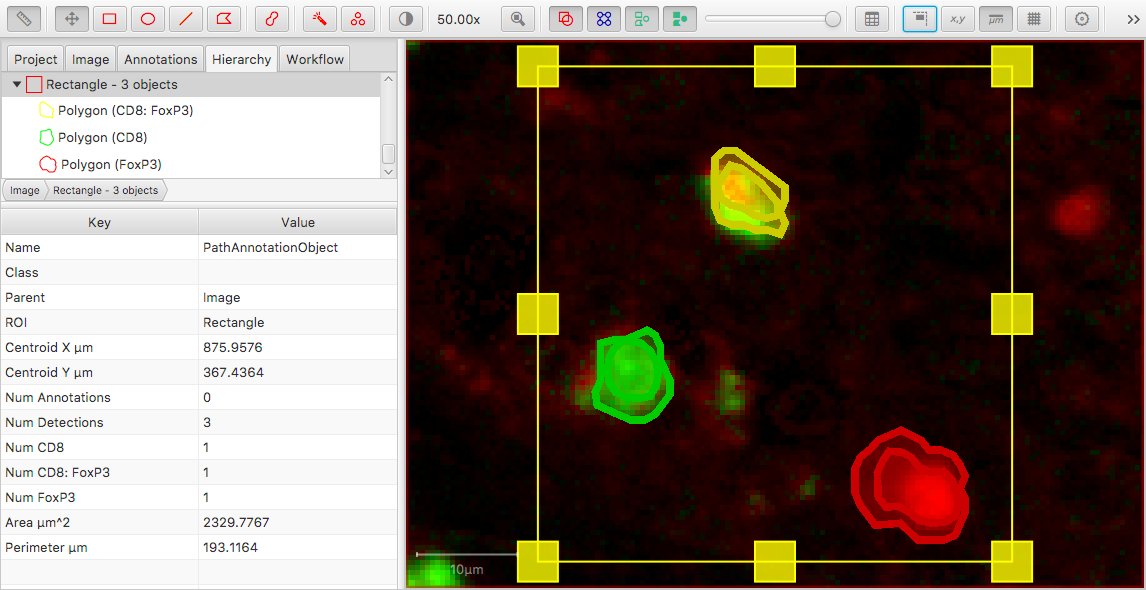
Note that the display of the objects was also changed here… in the Hierarchy view, only the last component of the classification was previously shown; now all parts are shown.
For & against
In favor of the change
Personally, I think the change is an improvement. It is more intuitive, and means that summing up all the cells with (non-base) classifications gives the ‘right’ answer for the total number of cells in the examples above – which wasn’t previously the case.
It also opens the door to working more easily with multiplex images as described in Part #1 immediately. There may still be a need for scripts to do specific tasks, at least until a more user-friendly interface can be developed, but the most important thing is that the data can be suitably represented, stored and queried. That’s a big step forward.
Against the change
Someone might potentially depend on the current behavior. They might be using sub-classifications for something other than intensities, and rely on everything remaining unchanged.
Having a measurement with the same name but different meaning would be a pretty mean thing to do for someone with a lot of custom scripts depending on this, or has simply calibrated their brain to interpret the numbers as they currently are.
Other options
A few alternatives that come to mind:
- Keep things as they are; multiple classifications could be supported some other way (e.g. with a helper class that contains the classified objects, rather than the classified objects containing their classifications). But this would need to be developed.
- Rather than reusing the same naming structure
Num Class, the measurement might be calledN ClassorCount ClassorClassified Class… or really anything that is totally different. By breaking everything that depends on the textNum, it draws attention to the fact there has been a change. - Rather than appending
(base)to the counts that include sub-classifications, append(only)(or something like that) to the counts that aren’t subclassifications. It would avoid the problem… but I think look ugly in perpetuity.
In conclusion
I tend towards making the change and clearly documenting it in the next QuPath release. I think the real benefits outweigh the possible risks. I would also think of bundling it up in a bigger change (e.g. v0.2.0) along with many other improvements that are on the way… such as switching to a new OpenCV version, updating dependencies and possibly even moving to Java 11.
But given how widely QuPath is now used, I don’t really want to unilaterally take that decision without explaining and having the possibility to discuss it. Also, even if it happens, there are also some details I’m not really decided on, such as the naming scheme ((base) or (parent) or something else?), and when it makes sense to count sub-classifications and when not.
Opinions and suggestions on all of this are welcome.
If nothing else, this may help give some insight into the kind of thing that keeps me up at night. This was all much easier before QuPath was open source, and when I made changes I was only breaking things for myself…