QuPath + Bio-Formats update
Improved image support (again) - including for 32-bit multichannel Vectra images
In the first post I mentioned a decision to leave my job in January. Since then, I’ve spent quite a bit of time travelling around, visiting various places and people, and deciding on the next step.
That decision has now been made and as of September I’ll be back in academia – this time as a PI, and with a continued focus on bioimage analysis and digital pathology. I’ll probably write a bit more about that some other day, once I’ve started the new position.
Anyhow, part of the travelling included attending the OME Annual Users Meeting for the first time, and being able to meet some of the fantastic team behind Bio-Formats. Since then I’ve been planning to revisit and revise the QuPath Bio-Formats extension.
I eventually got around to it last week, and a few of the improvements are particularly noteworthy.
Testing times
Firstly, I’ve finally set up a mechanism to test large numbers of images, checking if they can be read successfully with Bio-Formats + QuPath, and also to compare extracted metadata and pixel values with the built-in Bio-Formats + ImageJ combination.
Now, if I receive an image that fails in QuPath, but works with Bio-Formats elsewhere, it will be added to my test set and I’ll try to make sure that any fix doesn’t break something else.
With this in place, I started exploring the sample images at https://downloads.openmicroscopy.org - focussing on the pathology-related ones - to see what would fail. One collection did: the TIFFs in the ‘Vectra-QPTIFF’ directory, which the ReadMe.txt states were processed by inForm.
Further investigations and code updates has led to…
Support for 32-bit, multichannel images
The trouble turned out to be that the images were 32-bit floating point. These are now supported, opening up a whole new set of informatively colorful images for which QuPath can be used.
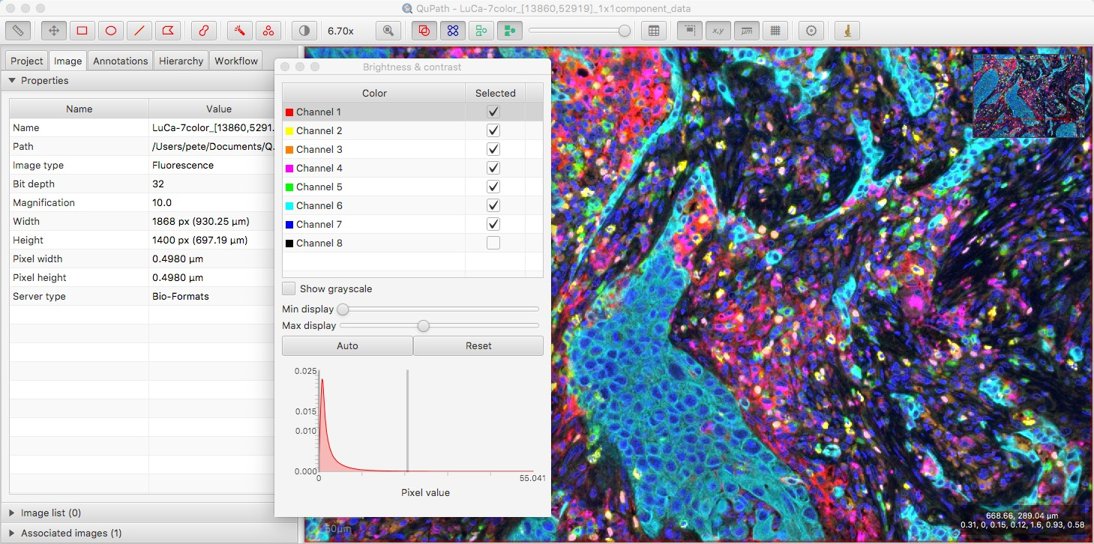
Original TIFF image LuCa-7color_[13860,52919]_1x1component_data.tif from Perkin Elmer, part of Bio-Formats sample data available under CC-BY 4.0).
While being able to view the image is a start, identifying more and more channels with the names Channel 1, Channel 2, Channel 3 etc. just won’t do.
Channel name information is available in Bio-Formats, and now this can be requested from a QuPath ImageServer with a getChannelName(int current) method. The following script shows it in action:
def server = getCurrentImageData().getServer()
for (c in 0..server.nChannels()-1)
print 'Channel ' + (c+1) + ': ' + server.getChannelName(c)
Better news is that with the latest v0.1.3 beta these names can now be seen using the Brightness/Contrast command:
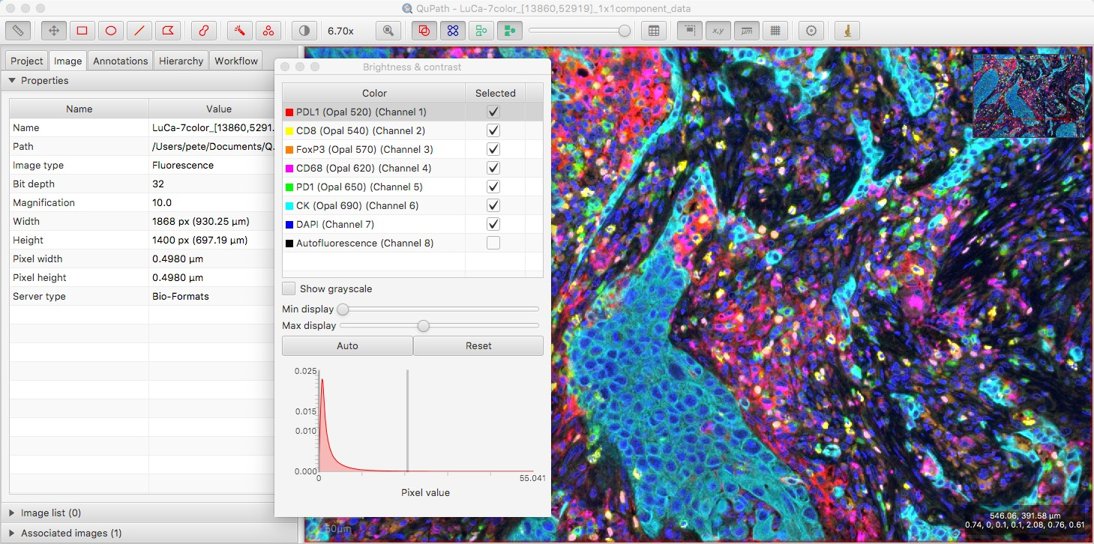
Other possibilities when working with such multichannel images will be the topic of a future blog post.
Developer note
To preserve backwards-compatibility,
getChannelName(int channel)doesn’t involve a change to theImageServerinterface, and not allImageServerimplementations will necessarily have the method. It is therefore requested within the Brightness/Contrast command using reflection.However, whenever QuPath undergoes a more substantial (not backwards-compatible) update, a method to request the channel name will be added.
This will take some more planning, as it should also include a method to change a wrong channel name, along with setting other incorrect metadata (which isn’t currently possible).
Improved metadata parsing
As I poked around some more inside Bio-Formats, and made further explorations of image file formats, I learned a few things that have resulted in some additional changes.
Choosing objectives
Firstly, @lacan pointed out complexities of which I wasn’t aware in determining the objective lens magnification. The previous method typically worked, but in the OME-XML used by Bio-Formats there’s the possibility of having different instruments and objectives, and if there is more than one then the process of identifying which belongs to which image isn’t entirely straightforward.
The extension now uses a new approach, which I hope will prove more robust.
Resolving resolutions
Secondly, anyone working with whole slide images knows they are stored as image pyramids, i.e. at different resolutions. It’s less straightforward than one might expect to figure out which resolutions are present.
QuPath requires a ‘downsample factor’ for each sub-resolution image. One straightforward method to determine this is to average the ratio of the width and height of the full-resolution image to each sub-resolution image. In OpenSlide, this calculation is done on QuPath’s behalf - and QuPath just reads these values.
Bio-Formats doesn’t provide the values directly itself, so they need to be calculated instead. It seemed logical for QuPath to use the same approach to that described above, but it doesn’t actually work in every case.
Weird things can happen with some file formats, such as horizontal and vertical ratios being wildly discordant; I’ve seen this with a few Olympus .vsi files.
Furthermore, using the ‘averaged ratio’ method, the calculated downsamples are often almost-but-not-quite what you might expect. For example, running the following script to print the downsamples
print getCurrentImageData().getServer().getPreferredDownsamples()
for the OpenSlide sample image CMU-2.svs results in the following:
[1.0, 4.000131319763625, 16.003678402522333, 32.01905559532393]
This indicates that there are 4 pyramid levels, with each downsample being tantalizingly close to a power of two.
Getting the downsample wrong by even a tiny amount can lead to problems… sometimes. The following screenshot shows the same image opened in QuPath with the previous Bio-Formats extension (v0.0.6, left) and with OpenSlide (right).
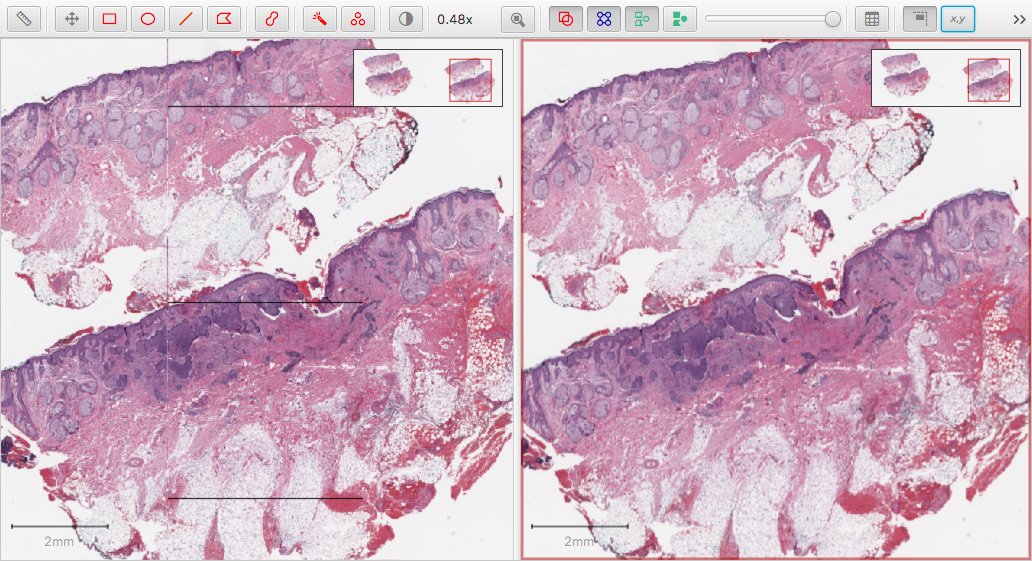
The square artefacts reveal that something is amiss – but it is only evident in certain regions of certain images when viewed at certain magnifications. And, as far as I could tell, the downsamples were the same in both cases (Bio-Formats and OpenSlide): yet clearly something different is going on somewhere.
This has been addressed in v0.0.7 by adapting the calculation of the downsamples when using Bio-Formats. The screenshot below shows the same image at the same magnification as before - this time opened using Bio-Formats, but without the artefacts.
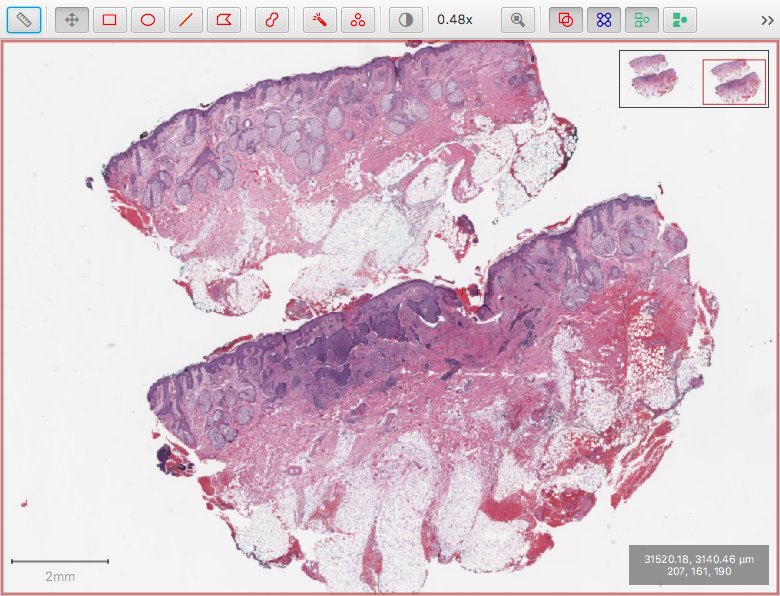
The crucial difference is in how the downsamples are calculated. The output of the script is as follows:
[1.0, 4.0, 16.0, 32.0, 76.25886983082707]
This reveals three things:
- the downsamples are now rounded whenever they are very close to being an integer value
- Bio-Formats actually accesses an additional pyramid level that OpenSlide does not provide (my guess is that OpenSlide considers this to be a thumbnail)
- it’s not safe to assume that the downsample factors will always be a power of two (indeed, some Zeiss
.cziimages seem to use powers of 3)
The logic behind the calculations is slightly involved (and .vsi still needs to be treated as a special case), but the relevant code is here.
I hope this will address the issues, but as is so often the case here, I only have a small number of images myself – and even fewer that I work with regularly – in a limited range of file formats. So it depends on feedback from users to find out if and when this may be wrong, and if a further refinement is needed.
This leads me to wonder whether a similar logic ought to also be applied when using OpenSlide, rather than using the precomputed downsample values given by OpenSlide itself… I haven’t done that yet, as the current approach does not seem obviously broken.
Viewing metadata
As described above, QuPath can use different libraries to read images - including OpenSlide and Bio-Formats. Generally, each library has its own way of representing metadata… some of which is then extracted and used by QuPath.
There are three places where the metadata can go wrong:
- the software writing the image in the first place
- the library attempting to read the metadata from the image
- QuPath’s effort to request and interpret the metadata from the library
Hopefully there won’t be too many problems at the third stage (and fewer still with the latest updates), but in cases where ‘the image won’t open’ or ‘the image looks weird’ it’s important to be able to trace back to try to decipher what has gone on and when. If it’s on the QuPath extension side, it’s something I’m more likely to be able to fix.
With that in mind, there’s a new dumpMetadata() method for most ImageServers, which (where implemented) provides a String representation of whatever metadata the reader knows about:
- For OpenSlide, this is currently a JSON-formatted map of key-value pairs
- For Bio-Formats, this is currently the OME-XML
QuPath doesn’t itself have any nice way to show XML, but many web browsers do. The following script is how I write the XML to a temporary file, and then open it in Chrome.
Using this brings me closer to seeing what the true contents of the file are, and through this deciphering where a problem might lie.
Checking version numbers
Finally, it can be hard to keep on track of what versions of extensions and libraries are being used.
Under Help → Installed extensions you can see the extensions listed, and this now also includes the version of the current Bio-Formats library (i.e. bioformats_package.jar).
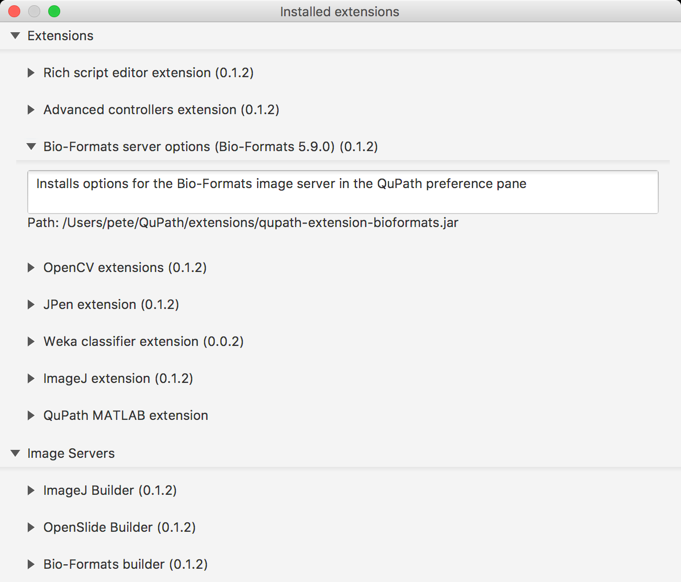
Note that this screenshot came from QuPath v0.1.2… which wrongly states that the Bio-Formats extension is also v0.1.2 (and unfortunate package-related confusion). The latest QuPath beta also fixes this, so that the correct Bio-Formats version (which here would be v0.0.7) is also shown.
Tip: If you double-click on the ‘Path’ part under an extension description, it should open up the directory containing the file… which is how I often access the QuPath extensions directory.