QuPath: Past, Present & Future
An overdue update on QuPath: What it is, where it's from & where it's going.
QuPath is open source bioimage analysis software, available at http://qupath.github.io.
If you’re reading this, there’s a good chance you’ve already encountered it.
One of the things that distinguishes QuPath from other open bioimage analysis tools (e.g. ImageJ, Fiji, CellProfiler, icy & ilastik) is its support for whole slide image analysis and digital pathology.
Broadly, if you find yourself working with ultra-large 2D brightfield or fluorescence images (perhaps 40 GB in size), potentially containing millions of cells that you want to be able to identify and classify, you might well want to try QuPath.
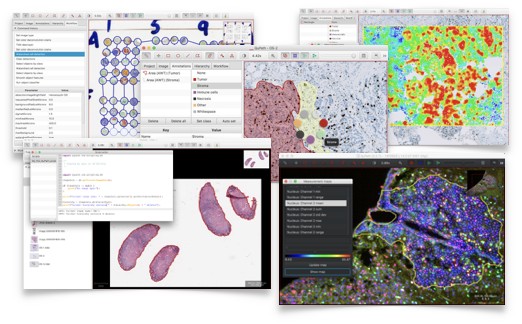
You might also want to try QuPath for other applications. QuPath doesn’t try to do everything, but for the things that it does do it tries to give lots of helpful features and shortcuts to make otherwise difficult tasks as painless as possible.
And because it’s open source, extensible and scriptable, if you’re computationally-minded and looking to develop new image analysis algorithms, then QuPath can give you a platform that takes care of a lot of the the essential-but-mundane tasks for you (e.g. handling large images, interactivity, visualization). This lets you focus on the most interesting parts of your work… and also the parts that lend themselves to being published in academic journals
What is this blog all about?
QuPath was released open source at the end of 2016. Since then the software has been downloaded thousands of times, referenced in various academic publications, taught in courses throughout Europe, and is being used in labs across the world.
However, after an initial flurry of updating activity, QuPath has not changed much since its first public incarnation.
The purpose of this post is to outline a little bit about the background to QuPath, describe where it is now, and give some personal reflections on where it’s going.
Disclaimer: All words and opinions are my own - and not those of any past or future employers. QuPath was developed at Queen’s University Belfast and released under the terms of the GPL v3.
The Past
Back from 2010-2012, I worked as an ‘image analysis specialist’ in the Nikon Imaging Center @ Heidelberg University, helping biologists analyze their imaging data.
This is where I really grew to appreciate the value of open source image analysis tools for research. They helped me enormously to work more efficiently.
What wasn’t always so efficient was trying to explain image analysis principles to lots of different people, especially when we spoke different languages (albeit using English words). I prefer to learn and explain things with visual aids - and ideally to choose my words at a pace slower than is really possible in real-time conversation.

So I ended up spending a long time trying to distill my explanations into a handbook:
Analyzing fluorescence microscopy images with ImageJ.
In the spirit of the open tools I was teaching, the handbook was made freely available too. Despite the regrettably long title, it became quite widely used. It now exists as a GitBook (especially handy for reading on mobile devices) and as a PDF.

Then in November 2012 I joined Queen’s University Belfast as a postdoc, and encountered digital pathology for the first time. Suddenly I was faced with a lot of large, unwieldy images that mortally offended my sense of right and wrong. To name just a few of their problems:
- they were far too big
- the staining wasn’t much good for quantification
- they were almost invariably JPEG-compressed
I also didn’t particularly like pink, and certainly wouldn’t have chosen to contrast it with purple (or seemingly, in some cases, a slightly darker shade of pink).
In time I came to appreciate the good sides of pathology images, and the exciting potential in the field. Still, I wanted to use my favorite open source software to handle the huge images… but try as I might, I didn’t find a good way to do so. There are great tools for huge 3D, 4D or even 5D datasets - but handling 2D is a bit different.
Based on the tools that I liked for microscopy data, I had an idea of what I wanted digital pathology software to do. One fateful day in late summer I started on the slippery slope of writing a whole slide image viewer - fully expecting to give up quickly. But through a sequence of felicitous (and sometimes nonfelicitous) events, it ended up becoming QuPath.
QuPath was designed to meet the goals of my postdoc project, but I hoped that it could be much more widely useful. And, of course, I hoped it could be open source.
QuPath was released towards the end of 2016. This came inside the final months of my time at Queen’s, and after I had accepted an offer to be industrious elsewhere. Hence the flurry of updates immediately after its release, while I was still at the university, followed by a sustained quiet period.
During the quiet period, QuPath nevertheless started to be discovered and used. A growing number of related papers have now been published (there is a list forming on the QuPath wiki).
The main paper describing the software - which I would ask you to please cite if you use QuPath in your work (since its future may well depend on it!) - is in Scientific Reports:
- Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). https://www.nature.com/articles/s41598-017-17204-5
The Present
After spending 2017 working in industry, I left in January this year.
My current plan is consciously and conspicuously vague; I’d want to argue ‘open’ is a better word. I left with the intention of returning to open research and science. Quite possibly academia. But before deciding anything too big, I plan to take a bit of time to read, think and learn new things. This also includes some teaching, and returning to QuPath.
It took a pretty huge amount of work to develop QuPath in the first place, and I’m happy that it’s now available and being used. But I’m much more excited about what it could do. Especially if more people get involved.
Because I am now not at the university where I developed QuPath, the situation is a bit different. Any work I do on it is voluntary. I still feel drawn to academia at heart, and during this mini career break remain interested in academic collaborations and publications.
But QuPath is sufficiently big, and my time sufficiently scarce, that I need to be very selective - with the priority being what I find really interesting or important. I’ve got a backlog of such ideas I’m itching to work on, and am trying (with limited success so far) to protect enough time to pursue these. I think they could also be particularly interesting for anyone using QuPath, and I’ll write more about them in due course.
Ongoing support and maintenance of the software remains a priority as well. For the past few weeks I’ve been making changes and updates on my own fork. I’ve already fixed most of the bugs and annoyances I know of in v0.1.2 (including the Windows-specific ones that had not previously bothered me personally).
I plan to continue working there for a while longer, before figuring out when/how to merge it all back together into the main software. Nevertheless, I’ll write another post soon about how to access these changes early, try them out, and give feedback.
The Future
Since I’m returning to QuPath after a hiatus, I’ve had some time to think about how to handle it. Even now, it’s a fairly big job to respond to the volume of questions and queries. I’m not complaining, and it’s great to know that it’s useful. Still, I’m looking for ways to help make the ongoing development sustainable.
This blog is part of it. I wanted a place for announcements and discussions, and to post scripts, descriptions, updates and other information about the project. I hope this might be helpful for anyone wanting to get involved, or to learn a bit more about how to make the most of the software.
Posts in the future will generally be shorter than this one, largely because I’d rather be coding stuff.
Another part involves teaching and meeting other users and developers.
In this regard, there will be a QuPath workshop at the EPFL in Switzerland from 17-18 April 2018. This will be a great chance to meet with anyone interested in QuPath who is able to join. There is more information at https://wiki.epfl.ch/qupath.
Anyhow, I hope that helps clarify what is going on. Now that this blog exists, I plan to use it to add updates more regularly over the next few months. After finally succumbing to Twitter, I might use that too.
In the meantime, if you’d like to discuss anything further, it shouldn’t be too hard to find ways to contact me. Or, even better, if you sign up for the EPFL workshop we can speak in person.