QuPath v0.2.0-m6 available
Sixth milestone on the path to QuPath v0.2.0
QuPath v0.2.0-m6 is now online!
This milestone contains two important bug fixes, one big change and several smaller improvements.
Warning! It’s generally not a good idea to switch between different QuPath versions. You should be able to read older data files within v0.2.0-m6, but the reverse might not be true. Backup often!
If you find bugs, please report them on GitHub or image.sc - don’t just stick with an older milestone that seemed to work!
For all other questions or discussions, please use image.sc.
Two important bug fixes
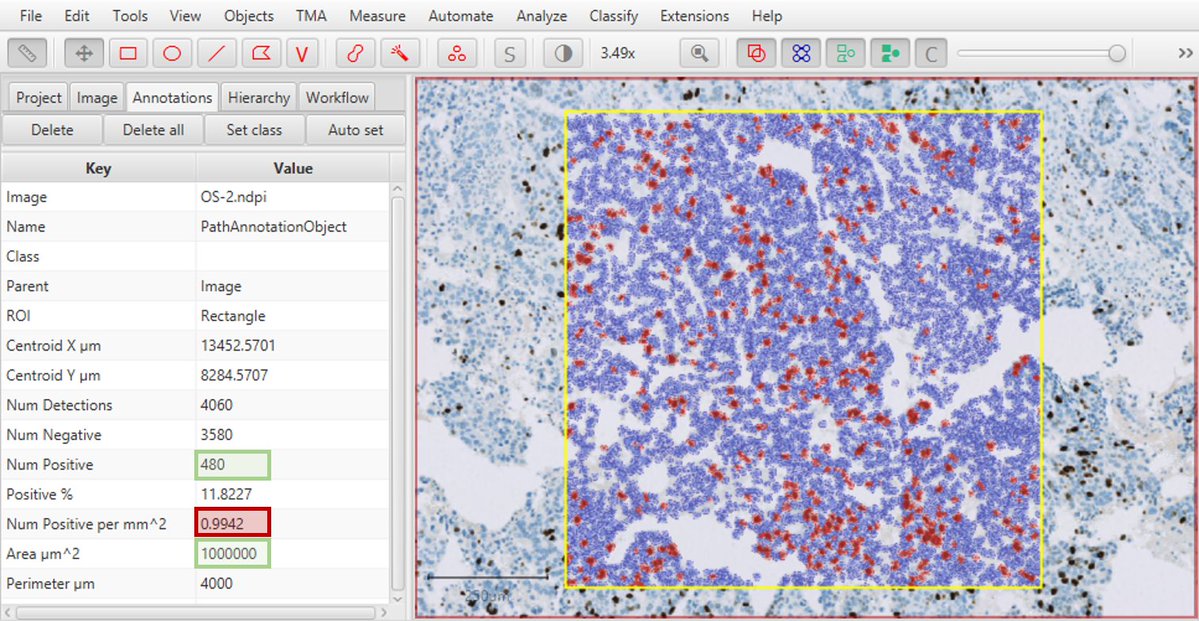
1) In v0.2.0-m5, the Positive per mm^2 measurement could be wrong. It would divide by the image area, not the annotation area - so to get the correct measurement you would need to calculate the value yourself based on Num positive and area. This bug only affected v0.2.0-m5, not other versions.
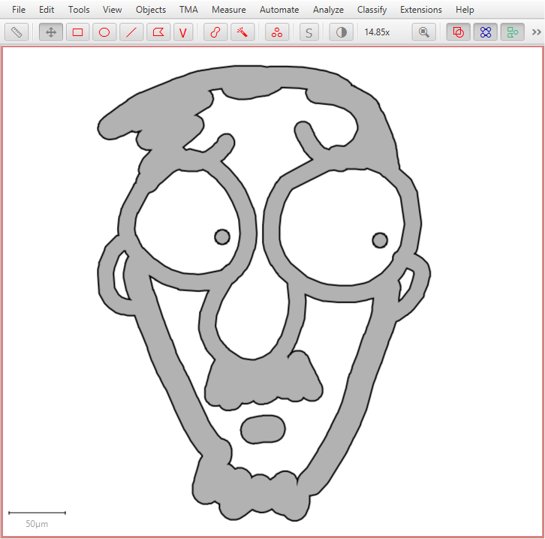
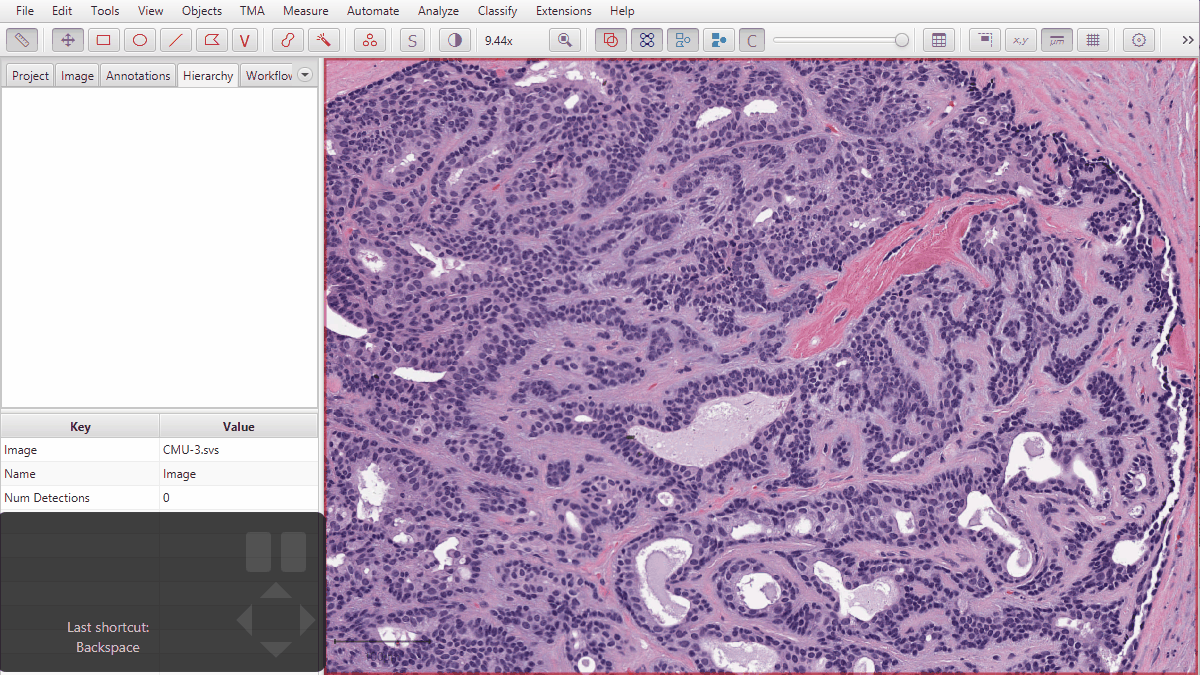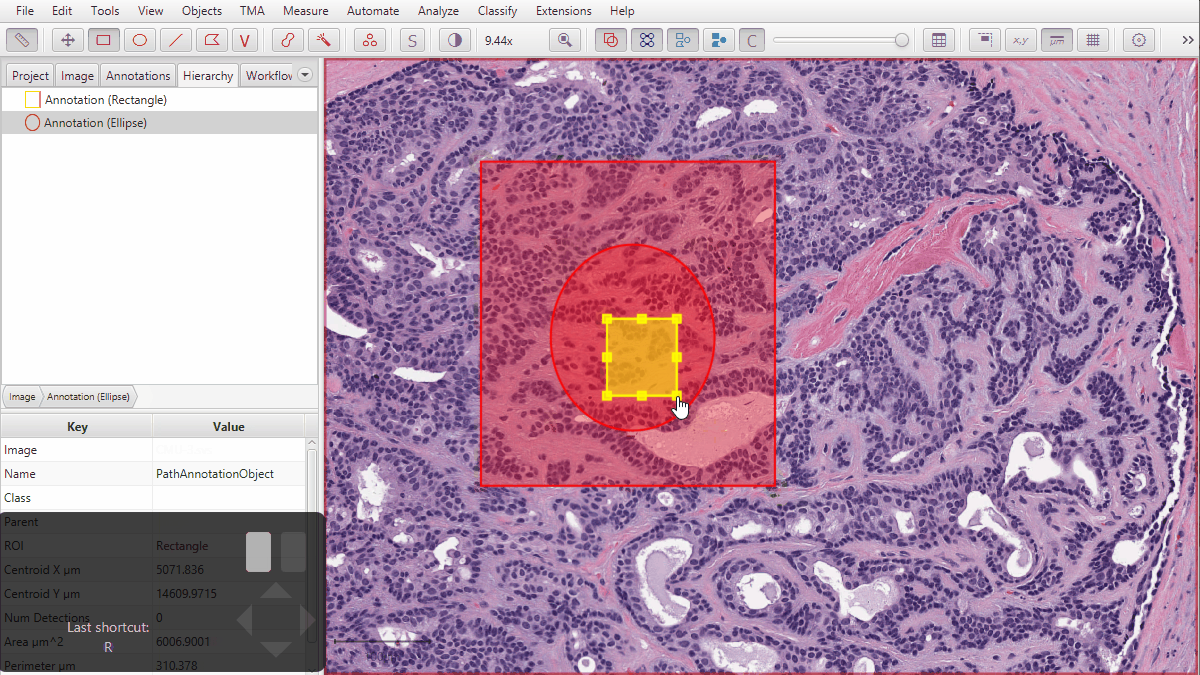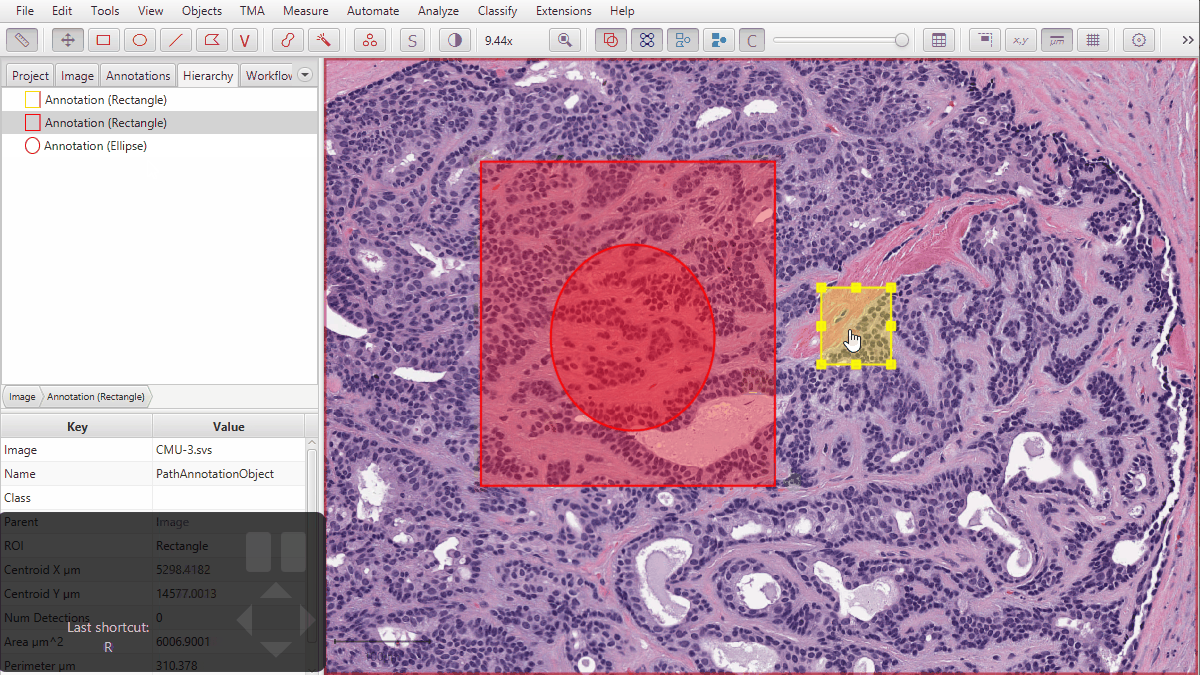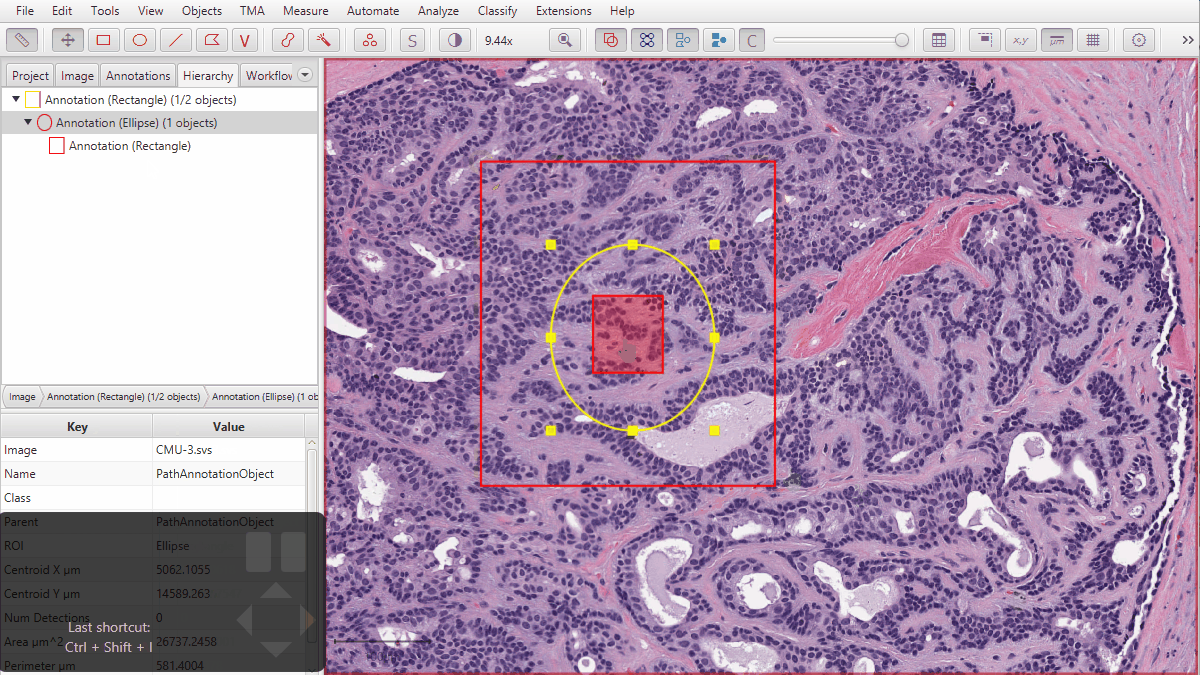
2) Converting ROIs with ‘extra pieces nested within holes’ into Java Topology Suite Geometries could lose the nested bits. Most ROIs aren’t like this, not all commands involve JTS, and not all JTS-related commands depend upon the conversion being perfect… so the impact of any error should be quite limited - but it has now been addressed.
An example of a troublesome ROI with pieces inside holes is shown below.
One big change: The object hierarchy
The change to the way objects are handled in QuPath is important enough to have its own blog post. I recommend reading it to understand what is different.
To summarize the outcome:
- The user now has more control over how and when the object hierarchy is used - the default behavior is different (and simpler) than it had been.
- Things that worked before should still work (if they don’t, please report the bug!), but you should be aware that the precise way in which they work may not be identical.
- Some operations, especially involving many objects/annotations, should be a lot faster than they were previously.
Everything else
Most other changes are concerned with stability and performance.
New ROI type (impacts data files!)
As part of addressing the JTS bug above, AreaROI has been replaced with GeometryROI.
What does that mean? Well, complex ROIs created in v0.2.0-m6 may not be readable in earlier QuPath versions. So please avoid modifying files in m6 if you plan to continue with an earlier QuPath version.
But it also means that some processing should be much faster, since there is less conversion between different ROI representions required.
To avoid losing work from the past, it should still be possible to read older data files into m6. But you may find the performance isn’t as good when doing this, because QuPath will occasionally need to convert old-style ROIs to new ones - which can be slow if they are very complex.
Smoother drawing
In addition to changing how the hierarchy operates, the way annotations are painted on screen has been updated. This should hopefully mean the drawing tools (e.g. brush) work more smoothly, even when many other objects are nearby.
If you see any weird visual effects it suggests I’ve got this wrong… but I’m not aware of any problems.
Reorganized menus
Some menus have been reorganised, and some old commands removed (OpenCV-based cell detection, local binary patterns) on the understanding they were rarely/never used and just added clutter.
If I’ve removed something you need we can discuss it. The reorganization shouldn’t really matter, because you use
Ctrl + Lto find commands - right?!
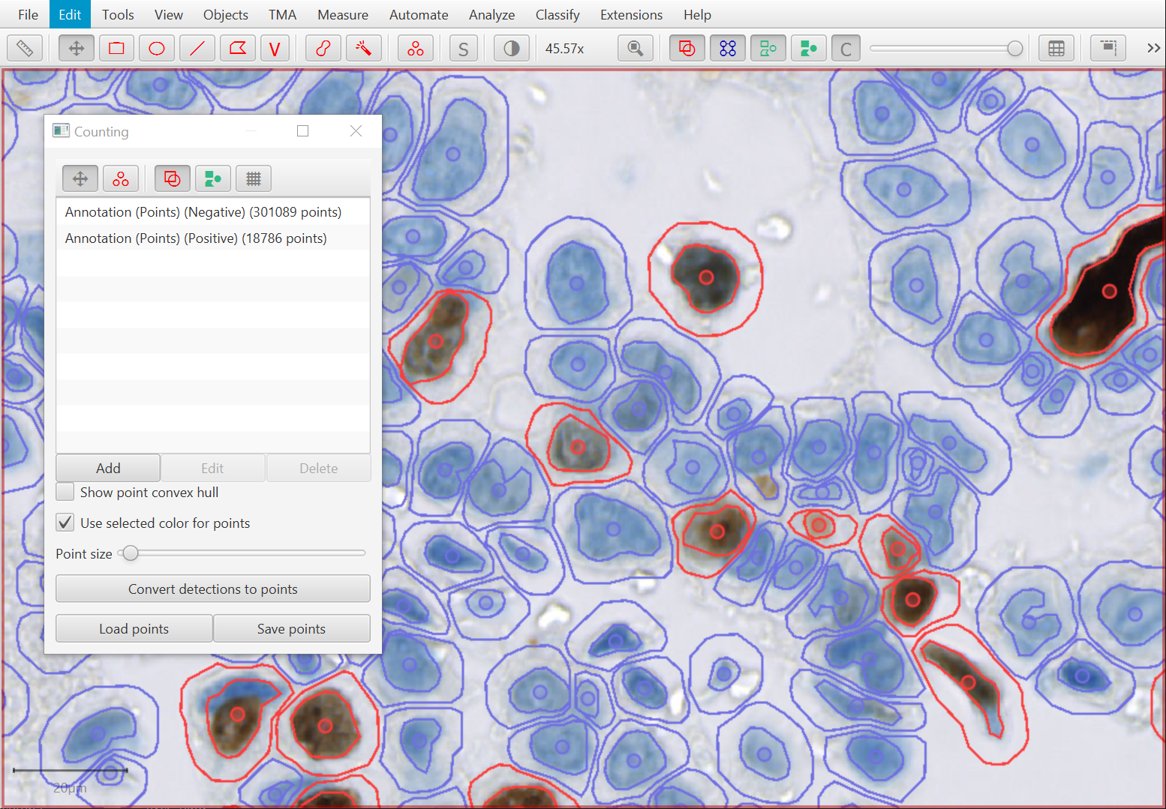
Create points from detections (and keep the detections)
The Convert detections to points button under the Counting tool now uses nucleus centroids (rather than cell centroids) where appropriate, and makes deleting the previous detections optional.
If you have a lot of point annotations, QuPath should handle them better and faster.
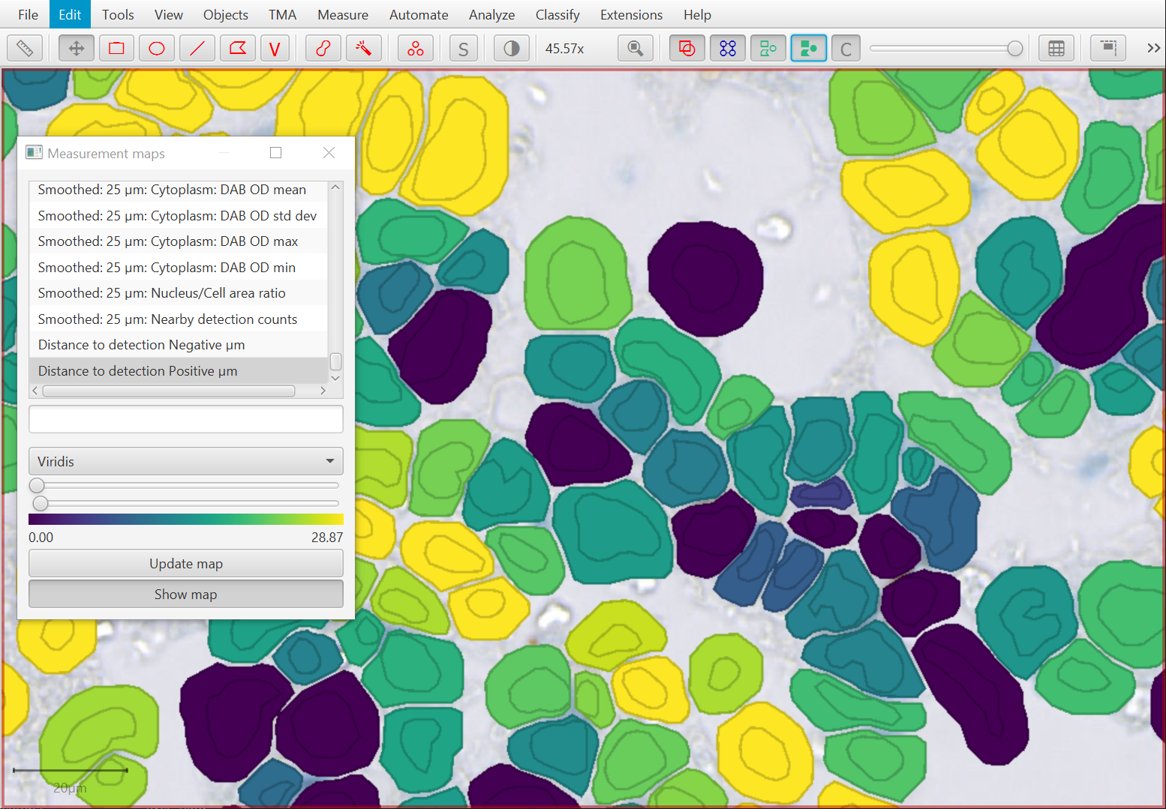
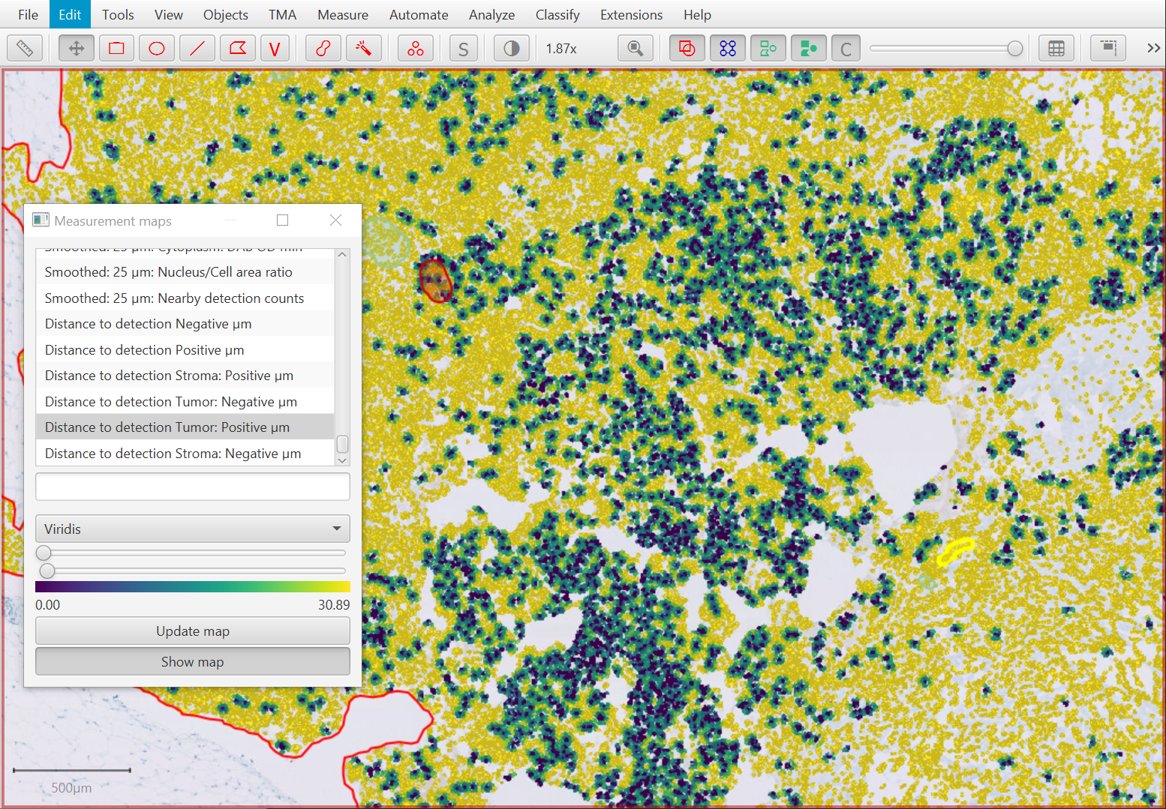
New Detection centroid distances 2D command
Building on the last option, there’s a new command that can be used to find distances to cells with different classifications.
Note there is a small rounding error… so that the distance from an object to itself is typically recorded as slightly greater than 0. This may be improved in a future release.
Other changes & fixes
- The pixel classifier shows live area measurements with ‘Classification’ output (in m5 this worked only with ‘Probability’ output)
- Extra shortcuts, including
Ctrl+Alt+Ato select annotations,Ctrl+Alt+Dto select detections - Undo/Redo and tile cache size information added to Memory Monitor
- Added support for ImageWriters to write to output streams
- Updated build script to Gradle 6.0
- Use bioformats_package.jar rather than separate dependences (easier to upgrade separately)
For full details on what has changed, see the commit log.