Script of the Day: Merging annotation results
Merge together the results from the previous script, to get one mega results table.
The previous script looked at exporting annotation results, with the results for each image being in a separate text file.
This script looks at combining these results into the same table. Most of it is just Groovy, and not specific to QuPath. Therefore this could also be done separately, outside QuPath, if necessary.
Problem
The previous script showed how to export annotation measurements for each individual image. But often it’s useful to have measurements for multiple images stored in the same table, with at least one column to identify the source image.
QuPath can’t automatically create such a table as standard within the main user interface. The main reason is that when you create a results table in QuPath (e.g. with Measure → Show annotation measurements) this is actually calculated dynamically - based upon all the objects currently in the object hierarchy.
One advantage of this is that the table can perform some updates after it has been created. For example, if I draw an annotation around some cells classified as positive and negative, the annotation results table will give the percentage of positive cells for that annotation. If I then move the annotation and release it somewhere else, the result updates to reflect the new annotation location.
However, because object hierarchies can include millions of objects, generating such dynamic tables across all the images in a project could (potentially at least) require a huge amount of memory and be very slow to generate. Therefore it is not possible, at least currently.
On the other hand, if you export the measurement table for an image as a text file, this basically gives a snapshot of the measurements in time. Only the value of the measurement is stored, not the underlying objects used to calculate it. Consequently, the files are usually very small (at least for annotations or TMAs, where we are typically dealing with hundreds of measurements at most). Combining these tables does not have the same memory or performance troubles.
There is, however, another complicating factor arising from the fact measurement tables are calculated dynamically. This is that the measurements (and therefore columns) can be different for each image. For example, one image might have cells with different intensity classifications (e.g. Tumor: Negative, Tumor: 1+, Tumor: 2+, Tumor: 3+), while perhaps another image only has cells classified as Tumor: Negative. In each case, the table will include only measurements that relate to classifications that are actually found within the image. So in the first example there would be more columns than the second.
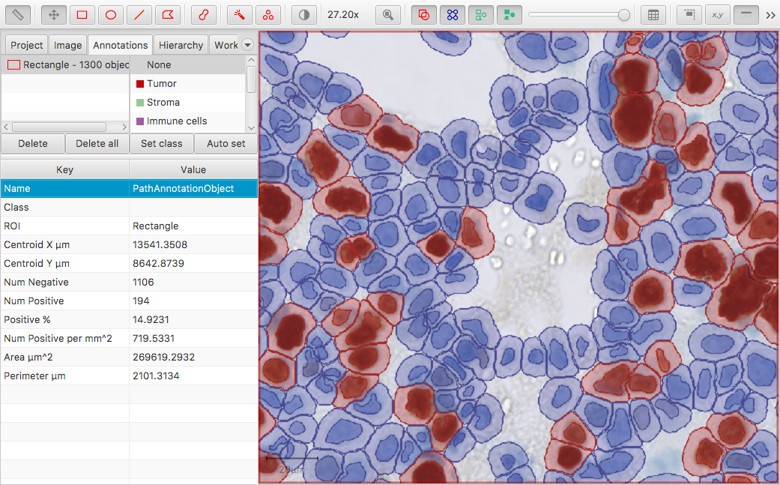
Dynamic measurements calculated when a single intensity threshold is applied to cells
Dynamic measurements calculated when three intensity thresholds are applied to cells
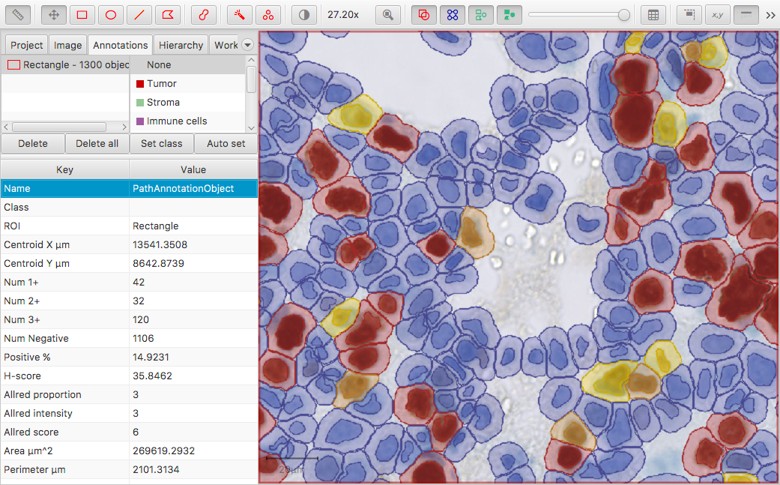
Again, this has advantages in normal use. For example, if you classify cells according to different staining intensities then QuPath will immediately recognize that you might want an H-score - and calculate it for you, without needing to be asked. But when it comes to merging tables, this feature can become troublesome - because we can’t count on each summary measurement table to contain the same columns.
Whether the advantages of dynamic measurement tables outweigh the disadvantages is up for debate - maybe the extra effort required to explicitly specify which measurements should be calculated would be worth it.
That is certainly something that might be considered in a future version of QuPath; for now, I am describing the situation as it is in v0.1.2.
Solution
To begin, let’s assume that results have already been exported to separate text files as described before, all in the same directory.
We then need to know where that directory is, and how to recognize the relevant files based on their extension. We also need to know how columns are separated, e.g. with a ‘tab’ mark or a comma. Because commas used to separate numeric data can be horribly problematic depending upon local language and formatting settings, the default is a ‘tab’ character.
The following lines specify the extension .txt and use of a tab separator, as well as the file name to be used for the output.
// Some parameters you might want to change...
String ext = '.txt' // File extension to search for
String delimiter = '\t' // Use tab-delimiter (this is for the *input*, not the output)
String outputName = 'Combined_results.txt' // Name to use for output; use .csv if you really want comma separators
We could specify the directory containing the files in a similar way - encoding the full path to the directory in the script, or generating it relative to the project (like in the export script).
However, another way is to show a directory chooser dialog to let the user interactively select the directory. Here, we use QuPath to create that prompt for us (this is really the only QuPath-specific bit of the script):
// Prompt for directory containing the results
import qupath.lib.gui.QuPathGUI
def dirResults = QuPathGUI.getSharedDialogHelper().promptForDirectory()
if (dirResults == null)
return
This makes use of a DialogHelper, which gives us a way to prompt the user for different kinds of file selections (single files, multiple files, directories…).
Note that it was necessary to import
QuPathGUIbefore we could use it to request theDialogHelper. We also check ifdirResults == null, which happens if the user cancels the dialog without choosing anything. In that case, the script ‘returns’, i.e. stops running.
Next, we need to get a list of all the relevant text files containing results, using the file extension defined above. Because we will write the output file to the same directory, we also need to make sure we don’t read it again - in case we run the script more than once.
There are lots of ways to do this with Java or Groovy. One of them is to create a new list of files, and then add to it any file within the directory that meets our required criteria, i.e. that it should be a file (and not a directory), its name should end with .txt (or .TXT - case shouldn’t matter), and it shouldn’t have the same name as the output.
// Get a list of all the files to merge
def files = []
for (file in dirResults.listFiles()) {
if (f.isFile() &&
f.toLowerCase().endsWith(ext) &&
f.getName() != outputName)
files.add(file)
}
This can be expressed a little more concisely in Groovy in the following way:
// Get a list of all the files to merge
def files = dirResults.listFiles({
File f -> f.isFile() &&
f.getName().toLowerCase().endsWith(ext) &&
f.getName() != outputName} as FileFilter)
Like above when specifying the directory, we need to prepare for the possibility that there aren’t any relevant files, and to stop gracefully with a meaningful message for the user. If there are relevant files, we can say how many and proceed.
if (files.size() <= 1) {
print 'At least two results files needed to merge!'
return
} else
print 'Will try to merge ' + files.size() + ' files'
Now it is almost time to start parsing the files, but there are a few things to consider first:
- To deal with the (potential) varying numbers of columns in the tables for each image, we can store each row of results as a
Map. This basically provides a ‘key, value pair’, of the format[key: value]. Here, the key is the name of the column, and the value is the value stored in the table. If we know the key, we can request the value later. - Because our table will have multiple rows, we need a
Listof such maps. We should also keep a record of all the column headings we ever see - so that we know when any of our maps is missing a value, and we can deal with that later by using a sensible default. - We also want to add an extra column for the file name - which isn’t stored in the results tables we are merging, but which will be needed later to recognize the image from which they came.
The following code sets this all up:
// Represent final results as a 'list of maps'
def results = new ArrayList<Map<String, String>>()
// Store all column names that we see - not all files necessarily have all columns
def allColumns = new LinkedHashSet<String>()
allColumns.add('File name')
Note that allColumns is defined as a LinkedHashSet. The Set part makes sure that every column name it stores is unique; if we add the same column multiple times, it still only remains once. The LinkedHash part indicates that it will be a set that maintains knowledge of the order in which entries were added (rather than sorting them alphabetically, or ordering them in some random way). This helps bring some sanity to the column ordering in the final output.
Now things progress rapidly, as we loop through all the results files doing the following:
- Read all the lines of text in the file
- Check if we have any data worth writing; if there is just one line, then that should be the header line
- Extract the first line (header), and split it according to the delimiter (tab) into column names
- Make sure we have all the columns in our
allColumnsset - For each row,
- Initialize a map to contain the results, with an extra first column giving the file name
- Add the values for each column to the map
- Perform a quick sanity check, to ensure there are not too many columns in any particular row (which would indicate something is wrong with the input or parsing)
- Add the map to the list of results
This is all accomplished in the following:
// Loop through the files
for (file in files) {
// Check if we have anything to read
def lines = file.readLines()
if (lines.size() <= 1) {
print 'No results found in ' + file
continue
}
// Get the header columns
def iter = lines.iterator()
def columns = iter.next().split(delimiter)
allColumns.addAll(columns)
// Create the entries
while (iter.hasNext()) {
def line = iter.next()
if (line.isEmpty())
continue
def map = ['File name': file.getName()]
def values = line.split(delimiter)
// Check if we have the expected number of columns
if (values.size() != columns.size()) {
print String.format('Number of entries (%d) does not match the number of columns (%d)!', columns.size(), values.size())
print('I will stop processing ' + file.getName())
break
}
// Store the results
for (int i = 0; i < columns.size(); i++)
map[columns[i]] = values[i]
results.add(map)
}
}
Now we’ve scooped up all the results, and it’s time to output them in an orderly way.
We have already defined our output file above. In general this should probably be tab-delimited (using the delimiter above) because of the localization troubles described above, but if anyone does wish to flagrantly disregard this advice and create a comma-separated file then we can sense than from the use of the .csv extension and adapt accordingly.
// Create a new results file - using a comma delimiter if the extension is csv
if (outputName.toLowerCase().endsWith('.csv'))
delimiter = ','
Finally, it’s time to write the output.
Writing files in Java can be slightly cumbersome (although it has improved over the years), involving creating streams or writers and then remembering to close them appropriately and handle any errors.
Groovy gives lots of potential shortcuts, one of which is used below:
int count = 0
fileResults.withPrintWriter {
def header = String.join(delimiter, allColumns)
it.println(header)
// Add each of the results, with blank columns for missing values
for (result in results) {
for (column in allColumns) {
it.print(result.getOrDefault(column, ''))
it.print(delimiter)
}
it.println()
count++
}
}
This essentially creates a PrintWriter, which is referred to within the {curly braces} as it (it’s possible to be more specific if you prefer). The above code also uses the String.join() method to help create and write all the column headings quickly, with the appropriate delimiter between them. After writing this as a line of text, the rest of the script loops through our ‘list of maps’, and then either writes the entry stored for the particular column name or else creates a blank column, with '', if there is no measurement available.
The reason for including a count was so that we can give a nice message at the end, indicating the extent of our achievement. Printing also helps let the user know that the script is finished:
print 'Done! ' + count + ' result(s) written to ' + fileResults.getAbsolutePath()
And that’s it!
The script is slightly long and involved, but it is also quite general - so can be reused across lots of projects.
Always being careful, of course, and checking that it does what is expected in every case. But that’s true for every script.